simdata: NORTA based simulation designs
Michael Kammer
2024-12-03
Source:vignettes/NORTA_demo.Rmd
NORTA_demo.RmdIntroduction
This document describes the workflow to define NORmal-To-Anything
(NORTA) based simulation designs using the simdata package.
The method is very useful to re-create existing datasets through a
parametric approximation for usage in simulation studies. It is also
quite easy to use, and allows the definition of presets for sharing
simulation setups. General details of the methodology and further
references are given in e.g. Cario and Nelson
(1997) and Ghosh and Henderson
(2003).
In this vignette we will prefix all relevant function calls by
:: to show the package which implements the function - this
is not necessary but only done for demonstration purposes.
Outline of NORTA
The goal of the NORTA procedure is to produce identically independently distributed (iid) samples from random variables with a given correlation structure (Pearson correlation matrix) and given marginal distributions, thereby e.g. approximating existing datasets.
Following Ghosh and Henderson (2003), we want to sample iid replicates of the random vector . Denote by the distribution functions (i.e. the marginal distributions) of the components of , and by the correlation matrix of . Then NORTA proceeds as follows:
- Generate multivariate standard normal random vectors (i.e mean 0, variance 1) with a correlation matrix .
- Compute the random vector via , where denotes the distribution function of the standard normal distribution, and is the quantile function of .
The resulting vector
then has the desired marginal distribution. To obtain the target
correlation structure
,
the correlation matrix
for the first step has to be chosen appropriately. This can be achieved
via solving univariable optimisation problems for each pair of variables
and
in
and is part of the simdata package.
Caveats of NORTA
The NORTA procedure has some known limitations, which may lead to
discrepancies between the target correlation structure and the
correlation structure obtained from the sampling process. These are,
however, partly alleviated when using existing datasets as templates, or
by special techniques within simdata.
- Not all combinations of given marginal distributions and target correlation are feasible by the nature of the variables. This is not an issue when an existing dataset is used as template, since that demonstrates that the combination exists.
- The optimisation procedure to obtain
may lead to a matrix which is not positive definite (since the
optimisation is only done for pairs of variables), and therefore not a
proper correlation matrix. To alleviate this, the
simdatapackage ensures positive definiteness by using the closest positive definite matrix instead. This may lead to discrepancies between the target correlation and the achieved correlation structure. - NORTA cannot reproduce non-linear relationships between variables. This may lead to issues for continuous variables and categorical variables with more than two categories when the goal is to faithfully re-create a real dataset that features non-linear relations.
- The optimisation procedure to obtain may take a while to compute when the number of variables increases. This is alleviated through the fact that this computation has to be done only a single time, during definition of the simulation design. All further simulation iterations only use the optimisation result and are therefore not subject to this issue.
- When applied to an existing dataset, NORTA relies on the estimation of the target correlation matrix and marginal distributions. More complex data (e.g. special marginal distributions, complex correlation structure) therefore requires more observations for an accurate representation.
Comparison to other methods
NORTA is well suited to re-create existing datasets through an
explicit parametric approximation. Similar methods exist, that achieve
this through other means. A particularly interesting alternative is the
generation of synthetic datasets using an approach closely related to
multiple imputation, and is implemented in e.g. the
synthpop R package (Nowok, Raab, and
Dibben (2016)). Its’ primary aim is to achieve confidentiality by
re-creating copies to be shared for existing, sensitive datasets.
In comparison, synthpop potentially offers more flexible
data generation than NORTA, thereby leading to a better approximation of
an original dataset. However, synthpop is also more opaque
than the explicit, user defined specification of correlation and
marginal distributions of NORTA. This also entails that
synthpop can be generally used more like a black-box
approach, which requires little user input, but is also less transparent
than the manual curation of the simulation setup in NORTA. Furthermore,
NORTA allows easy changes to the design to obtain a wide variety of
study designs from a single template dataset, whereas
synthpop is more targeted at re-creating the original
dataset. Both methods therefore have their distinct usecases and
complement each other.
Workflow in simdata
Given the outline of the method, all the user has to specify to define a NORTA design on variables are
- A target correlation matrix
- The marginal distributions for each variable, given as quantile functions
These can be estimated from existing datasets of interest.
simdata offers a helper function to automate this process,
but the user can also specify the required input manually. We
demonstrate both use cases in the example
below.
Quantile functions for some common distributions
The required marginal distributions are given as quantile functions.
R provides implementations of many standard distributions which can be
directly used, see the help on distributions. The quantile
functions use the prefix “q”, as in e.g. qnorm or
qbinom. Further implementations can be found in the
packages extraDistr, actuar and many others
(see https://CRAN.R-project.org/view=Distributions).
Example
In this example we will setup a NORTA based simulation design for a
dataset extracted from the National Health And Nutrition Examination
Survey (NHANES), accessible in R via several packages (we use the
NHANES package in this demo).
Load dataset
First we will load the dataset and extract several variables of
interest, namely gender (‘Gender’), age (‘Age’), race (‘Race’), weight
(‘Weight’), bmi (‘BMI’), systolic (‘BPsys’) and diastolic blood pressure
(‘BPdia’). These variabes demonstrate several different kinds of
distributions. For a detailed description of the data, please see the
documentation at https://www.cdc.gov/nchs/nhanes.htm and for the
NHANES R package on CRAN. Here we are not concerned with
the exact codings of the variables, so we will remove labels to factor
variables and work with numeric codes. Further, we will only use data
from a single survey round to keep the dataset small.
df = NHANES::NHANES %>%
filter(SurveyYr == "2011_12") %>%
select(Gender, Age, Race = Race1, Weight,
BMI, BPsys = BPSys1, BPdia = BPDia1) %>%
filter(complete.cases(.)) %>%
filter(Age > 18) %>%
mutate(Gender = if_else(Gender == "male", 1, 2),
Race = as.numeric(Race))
print(head(df))## # A tibble: 6 × 7
## Gender Age Race Weight BMI BPsys BPdia
## <dbl> <int> <dbl> <dbl> <dbl> <int> <int>
## 1 2 43 1 98.6 33.3 100 70
## 2 1 80 4 85.9 28.5 124 72
## 3 1 35 4 89 27.9 108 62
## 4 1 57 4 96.9 28 112 70
## 5 1 57 4 96.9 28 112 70
## 6 1 57 4 96.9 28 112 70Estimate target correlation
Using this dataset, we first define the target correlation
cor_target and plot it.
cor_target = cor(df)
ggcorrplot::ggcorrplot(cor_target, lab = TRUE)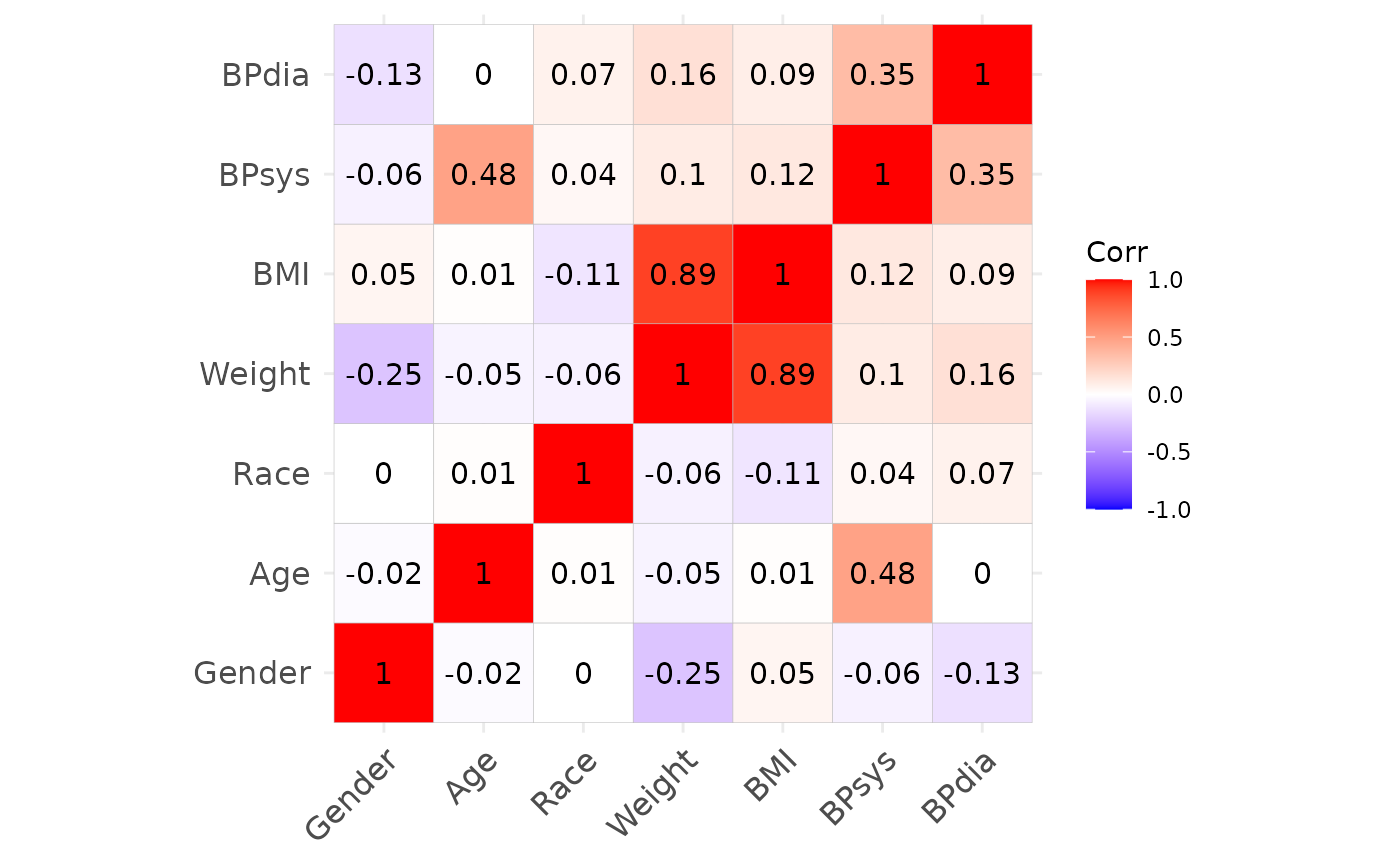
Define marginal distributions
Further, we define a list of marginal distributions dist
representing the individual variables. Each entry of the list must be a
function in one argument, defining the quantile function of the
variable. The order of the entries must correspond to the order in the
target correlation cor_target.
Automatically
simdata offers the helper function
simdata::quantile_functions_from_data() to automate
estimation of quantile functions from the available data. It does so
non-parametrically and implements two approaches, one more suited for
categorical data, and the other more suited to continuous data. In
practice the parameter n_small can be used to determine a
number of unique values required to use the latter approach, rather than
the former. See the documentation for more details.
dist_auto = quantile_functions_from_data(df, n_small = 15) Manually
We use the fitdistrplus::fitdist function to find
appropriate distribution candidates and fit their parameters. Decisions
regarding the fit of a distribution can be made using e.g. the Akaike
information criterion (AIC) or Bayesian information criterion (BIC)
displayed by the summary of the fit object returned by the function (the
lower their values, the better the fit).
In case a parametric distribution doesn’t fit very well, we instead make use of a density estimate and use this to define the marginal quantile function.
- Gender: a binomial distribution with .
- Age: the distribution is not very “nice”.
- We approximate it using a kernel density estimate using the
stats::densityfunction. - Note that the boundaries of the distribution can be more or less
smoothed with the
cutparameter. - To obtain a quantile function, first we integrate the density,
normalize it, and then use
stats::approxfunto derive a univariable quantile function.
- We approximate it using a kernel density estimate using the
- Race: a categorical distribution with 5 categories specified by
probabilities
- Frequencies of categories were identified by a simple call to
table()on the full dataset. - Can also be implemented using the categorical distribution from the
package
LaplacesDemonimplemented viaqcat
- Frequencies of categories were identified by a simple call to
- Weight: gamma distribution parameters estimated using
fitdistrplus::fitdist - BMI and systolic blood pressure: log-normal distribution parameters
estimated using
fitdistrplus::fitdist - Diastolic blood pressure: normal distribution parameters estimated
using
fitdistrplus::fitdistafter removing zero values from the data
The code to implement these marginal distributions is shown below.
dist = list()
# gender
dist[["Gender"]] = function(x) qbinom(x, size = 1, prob = 0.5)
# age
dens = stats::density(df$Age, cut = 1) # cut defines how to deal with boundaries
# integrate
int_dens = cbind(Age = dens$x, cdf = cumsum(dens$y))
# normalize to obtain cumulative distribution function
int_dens[, "cdf"] = int_dens[, "cdf"] / max(int_dens[, "cdf"])
# derive quantile function
# outside the defined domain retun minimum and maximum age, respectively
dist[["Age"]] = stats::approxfun(int_dens[, "cdf"], int_dens[, "Age"],
yleft = min(int_dens[, "Age"]),
yright = max(int_dens[, "Age"]))
# race
dist[["Race"]] = function(x)
cut(x, breaks = c(0, 0.112, 0.177, 0.253, 0.919, 1),
labels = 1:5)
# weight
fit = fitdistrplus::fitdist(as.numeric(df$Weight), "gamma")
summary(fit)
dist[["Weight"]] = function(x) qgamma(x, shape = 16.5031110, rate = 0.2015375)
# bmi
fit = fitdistrplus::fitdist(as.numeric(df$BMI), "lnorm")
summary(fit)
dist[["BMI"]] = function(x) qlnorm(x, meanlog = 3.3283118, sdlog = 0.2153347)
# systolic blood pressure
fit = fitdistrplus::fitdist(as.numeric(df$BPsys), "lnorm")
summary(fit)
dist[["BPsys"]] = function(x) qlnorm(x, meanlog = 4.796213, sdlog = 0.135271)
# diastolic blood pressure
fit = fitdistrplus::fitdist(as.numeric(df %>%
filter(BPdia > 0) %>%
pull(BPdia)), "norm")
summary(fit)
dist[["BPdia"]] = function(x) qnorm(x, mean = 71.75758, sd = 11.36352)What to use?
Both, the automatic and the manual way to specify marginals may be useful. The automatic way works non-parametrically which may be useful when a real dataset should be re-created, while the manual way allows to specify marginals parametrically which may be useful when the data is defined from purely theoretical specifications.
Simulate data
Now we can use simdata::simdesign_norta to obtain
designs using both the manual and automated marginal specifications.
After that, we simulate datasets of the same size as the original data
set using simdata::simulate_data, and compare the resulting
summary statistics and correlation structures.
# use automated specification
dsgn_auto = simdata::simdesign_norta(cor_target_final = cor_target,
dist = dist_auto,
transform_initial = data.frame,
names_final = names(dist),
seed_initial = 1)
simdf_auto = simdata::simulate_data(dsgn_auto, nrow(df), seed = 2)
# use manual specification
dsgn = simdata::simdesign_norta(cor_target_final = cor_target,
dist = dist,
transform_initial = data.frame,
names_final = names(dist),
seed_initial = 1)
simdf = simdata::simulate_data(dsgn, nrow(df), seed = 2)Results
Summary statistics of the original and simulated datasets.
summary(df)## Gender Age Race Weight
## Min. :1.000 Min. :19.00 Min. :1.000 Min. : 39.30
## 1st Qu.:1.000 1st Qu.:32.00 1st Qu.:3.000 1st Qu.: 66.58
## Median :1.000 Median :46.00 Median :4.000 Median : 79.40
## Mean :1.494 Mean :46.97 Mean :3.539 Mean : 81.88
## 3rd Qu.:2.000 3rd Qu.:60.00 3rd Qu.:4.000 3rd Qu.: 93.20
## Max. :2.000 Max. :80.00 Max. :5.000 Max. :198.70
## BMI BPsys BPdia
## Min. :15.70 Min. : 74.0 Min. : 0.00
## 1st Qu.:23.80 1st Qu.:110.0 1st Qu.: 64.00
## Median :27.60 Median :120.0 Median : 72.00
## Mean :28.57 Mean :122.2 Mean : 71.63
## 3rd Qu.:31.90 3rd Qu.:132.0 3rd Qu.: 78.00
## Max. :80.60 Max. :224.0 Max. :110.00
summary(simdf_auto)## Gender Age Race Weight
## Min. :1.000 Min. :19.00 Min. :1.000 Min. : 39.30
## 1st Qu.:1.000 1st Qu.:32.00 1st Qu.:3.000 1st Qu.: 65.50
## Median :1.000 Median :46.00 Median :4.000 Median : 78.90
## Mean :1.499 Mean :46.72 Mean :3.522 Mean : 81.04
## 3rd Qu.:2.000 3rd Qu.:60.00 3rd Qu.:4.000 3rd Qu.: 92.49
## Max. :2.000 Max. :80.00 Max. :5.000 Max. :143.60
## BMI BPsys BPdia
## Min. :15.70 Min. : 74.0 Min. : 0.00
## 1st Qu.:23.60 1st Qu.:112.0 1st Qu.: 64.00
## Median :27.30 Median :120.0 Median : 72.00
## Mean :28.34 Mean :122.0 Mean : 71.31
## 3rd Qu.:31.90 3rd Qu.:132.0 3rd Qu.: 78.00
## Max. :48.23 Max. :174.6 Max. :102.00
summary(simdf)## Gender Age Race Weight
## Min. :0.0000 Min. :16.01 Min. :1.000 Min. : 27.56
## 1st Qu.:0.0000 1st Qu.:32.10 1st Qu.:4.000 1st Qu.: 66.90
## Median :1.0000 Median :46.05 Median :4.000 Median : 79.85
## Mean :0.5098 Mean :46.85 Mean :3.542 Mean : 81.75
## 3rd Qu.:1.0000 3rd Qu.:60.21 3rd Qu.:4.000 3rd Qu.: 94.44
## Max. :1.0000 Max. :82.91 Max. :5.000 Max. :175.01
## BMI BPsys BPdia
## Min. :12.57 Min. : 67.03 Min. : 34.98
## 1st Qu.:23.90 1st Qu.:111.23 1st Qu.: 64.42
## Median :27.67 Median :121.13 Median : 71.99
## Mean :28.53 Mean :122.61 Mean : 72.00
## 3rd Qu.:32.44 3rd Qu.:132.88 3rd Qu.: 79.73
## Max. :56.66 Max. :189.50 Max. :115.30Correlation structures of the original and simulated datasets.
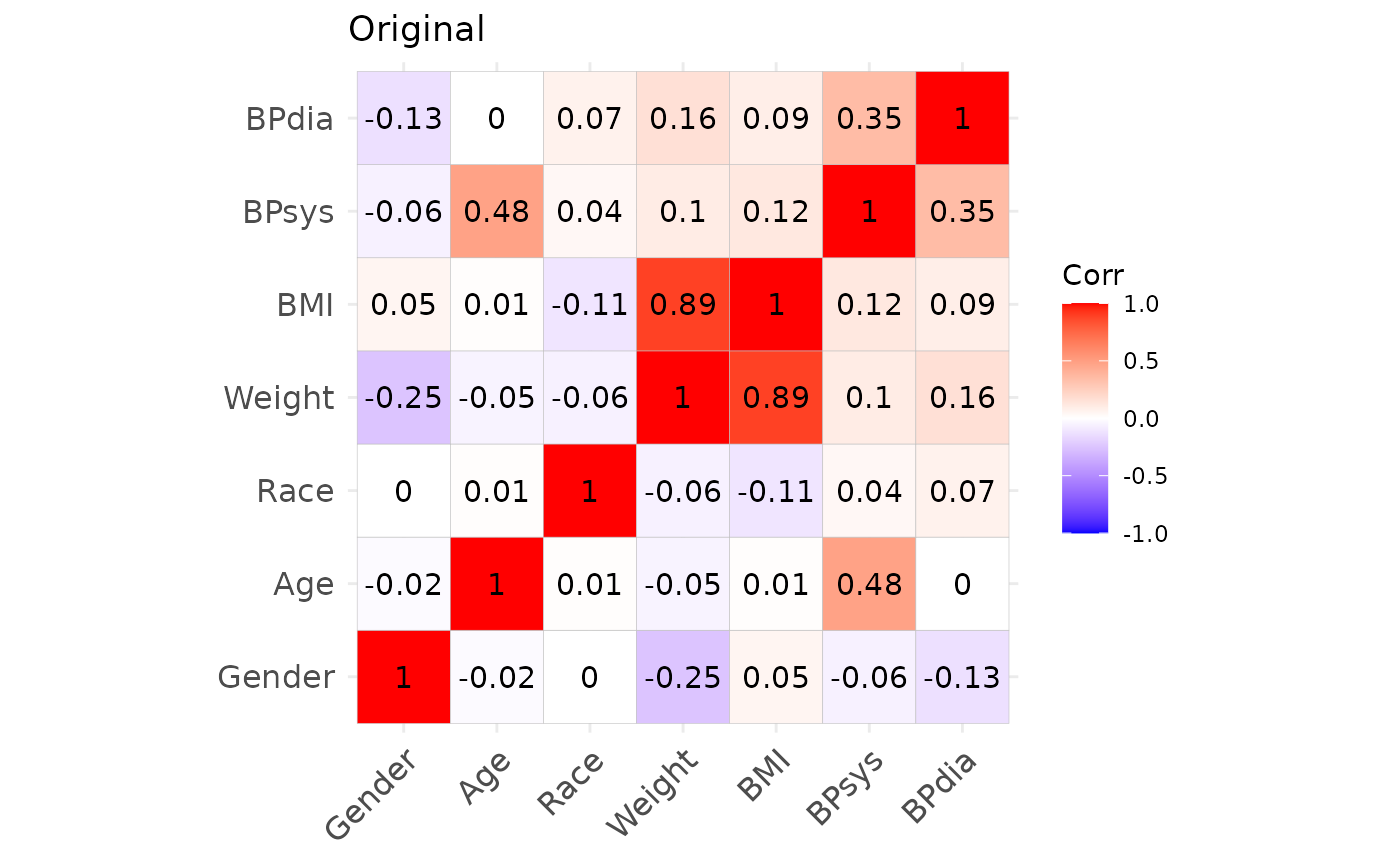
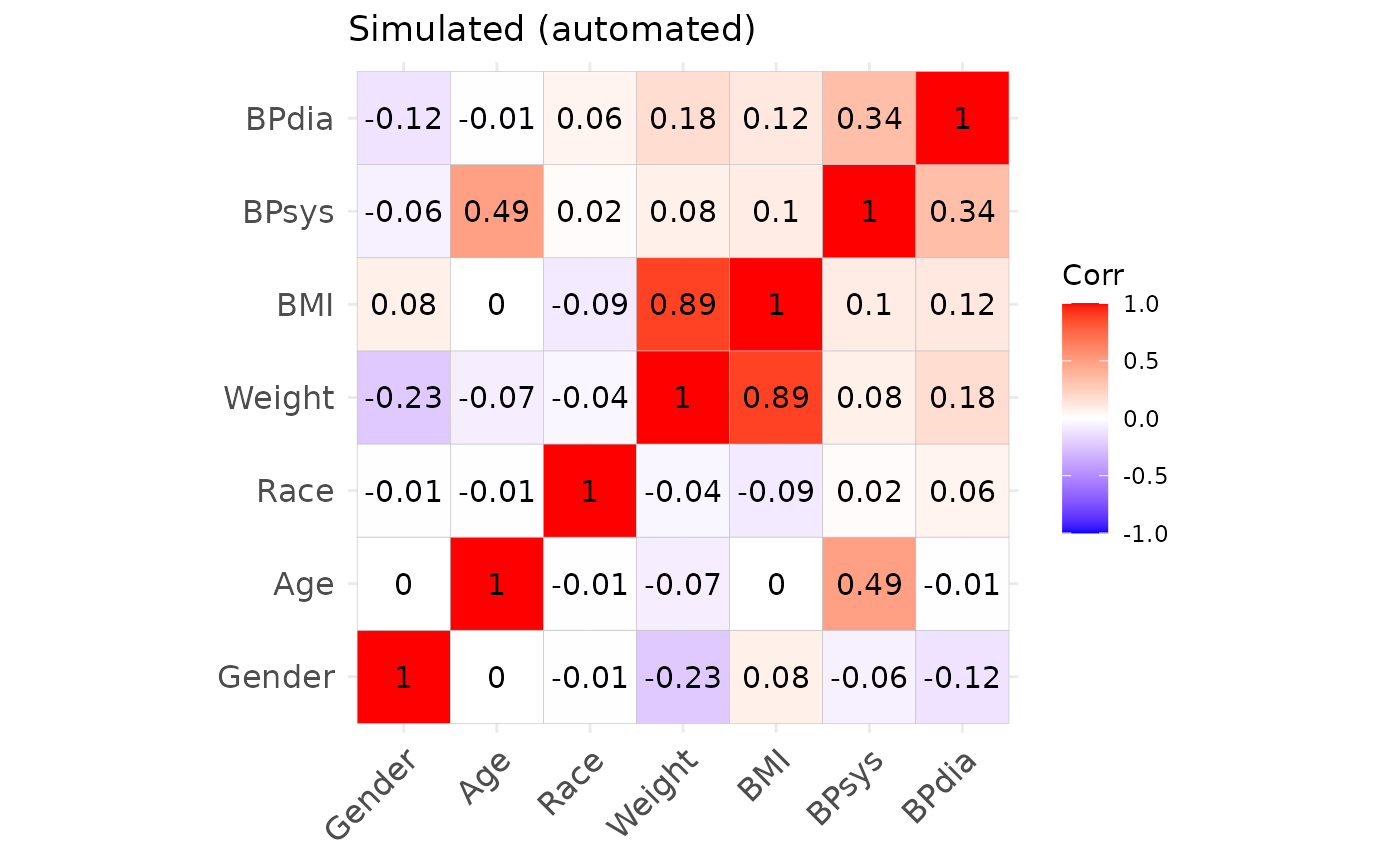
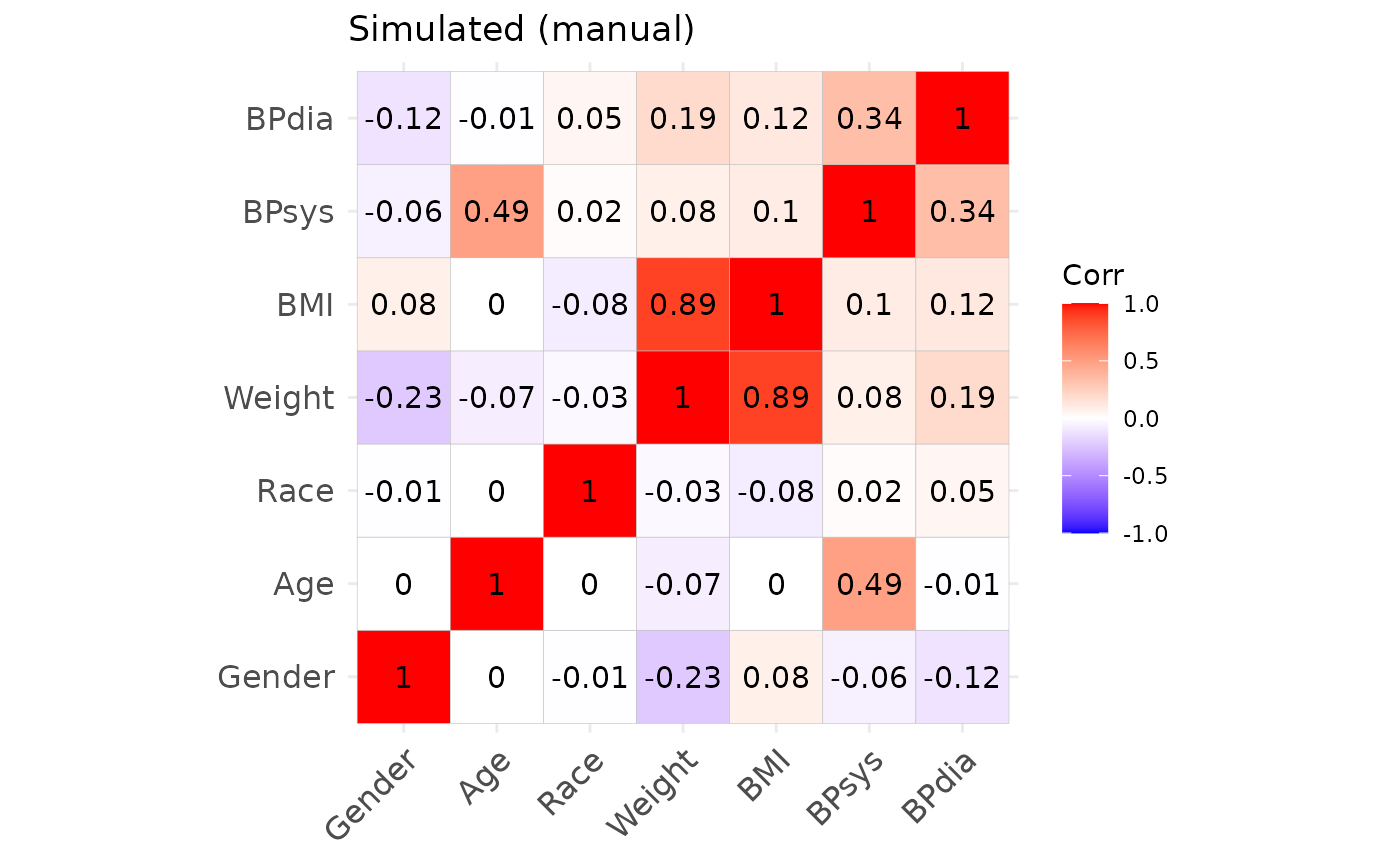
We may also inspect the continuous variables regarding their univariate and bivariate distributions. The original data is shown in black, the simulated data is shown in red. (Note that we only use the first 1000 observations to speed up the plotting.)
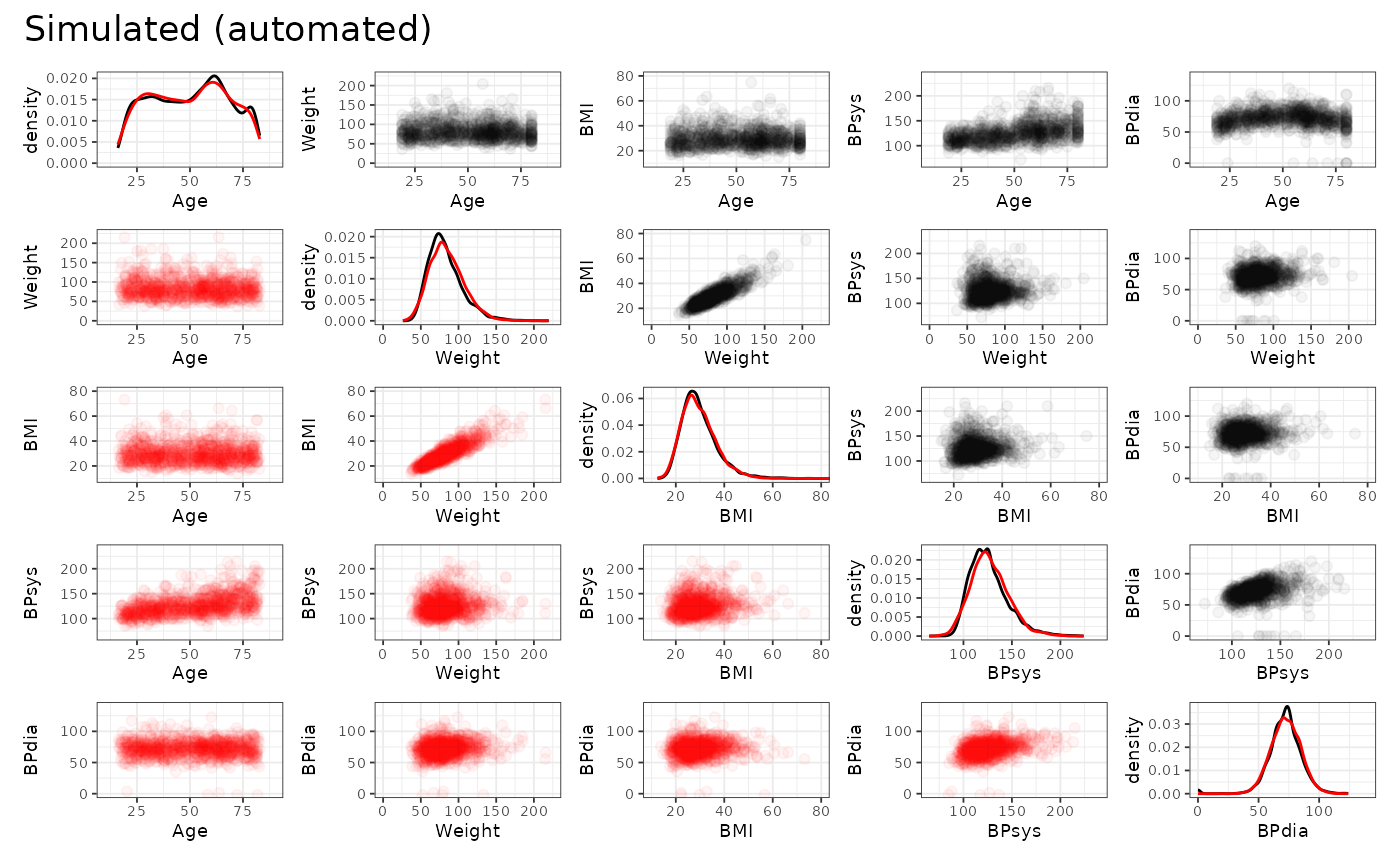
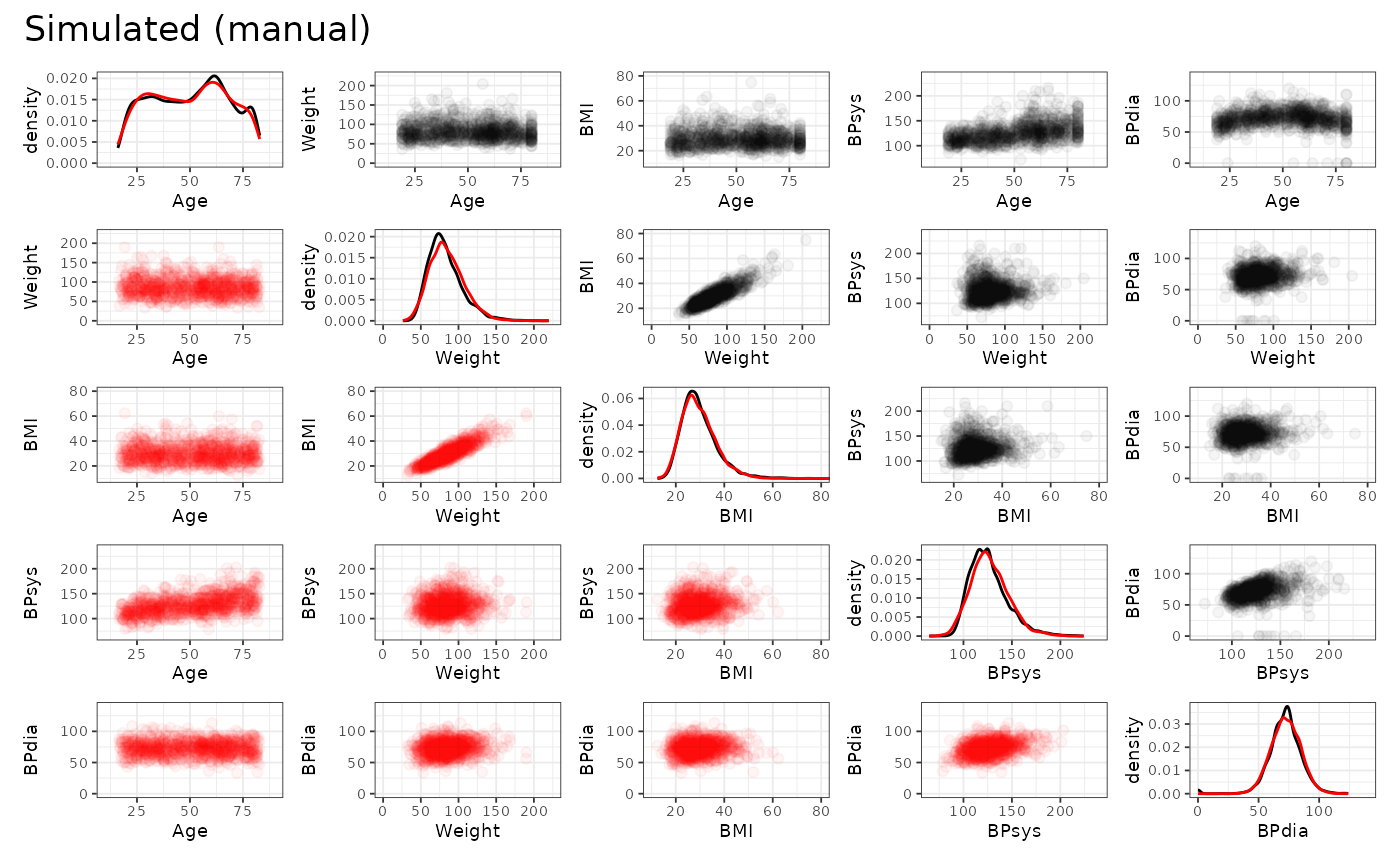
From this we can observe, that the agreement between the original data and the simulated data is generally quite good. Both, automated and manual specification work equally well for this dataset. Note, however, that e.g. the slightly non-linear relationship between age and diastolic blood pressure cannot be fully captured by the approach, as expected. Furthermore, the original data shows some outliers, which are also not reproducible due to the parametric nature of the NORTA procedure.
R session information
## R version 4.4.2 (2024-10-31)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 22.04.5 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
##
## locale:
## [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
## [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
## [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
##
## time zone: UTC
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggcorrplot_0.1.4.1 patchwork_1.3.0 ggplot2_3.5.1 dplyr_1.1.4
## [5] fitdistrplus_1.2-1 survival_3.7-0 MASS_7.3-61 simdata_0.4.1
##
## loaded via a namespace (and not attached):
## [1] sass_0.4.9 utf8_1.2.4 generics_0.1.3 NHANES_2.1.0
## [5] stringi_1.8.4 lattice_0.22-6 digest_0.6.37 magrittr_2.0.3
## [9] evaluate_1.0.1 grid_4.4.2 mvtnorm_1.3-2 fastmap_1.2.0
## [13] plyr_1.8.9 jsonlite_1.8.9 Matrix_1.7-1 fansi_1.0.6
## [17] scales_1.3.0 textshaping_0.4.0 jquerylib_0.1.4 cli_3.6.3
## [21] rlang_1.1.4 munsell_0.5.1 splines_4.4.2 withr_3.0.2
## [25] cachem_1.1.0 yaml_2.3.10 tools_4.4.2 reshape2_1.4.4
## [29] colorspace_2.1-1 vctrs_0.6.5 R6_2.5.1 lifecycle_1.0.4
## [33] stringr_1.5.1 fs_1.6.5 ragg_1.3.3 pkgconfig_2.0.3
## [37] desc_1.4.3 pkgdown_2.1.1 pillar_1.9.0 bslib_0.8.0
## [41] gtable_0.3.6 glue_1.8.0 Rcpp_1.0.13-1 systemfonts_1.1.0
## [45] xfun_0.49 tibble_3.2.1 tidyselect_1.2.1 knitr_1.49
## [49] farver_2.1.2 htmltools_0.5.8.1 labeling_0.4.3 rmarkdown_2.29
## [53] compiler_4.4.2